HW Polar Vortex & Machine Learning¶
Individual Work¶
Credits: 10%¶
Grading:¶
Yu-Chiao Liang (10%)
Deadline: xxx¶
The Tasks¶
Use zonal winds (U) at 10 hPa to categorize different type of stratospheric polar vortex.
Understand the basic idea of unsupervised machine learning technique.
Be familiar with using the hierarchical clustering with Python package - scipy.
Use other clustering method, for example, k-means cluster.
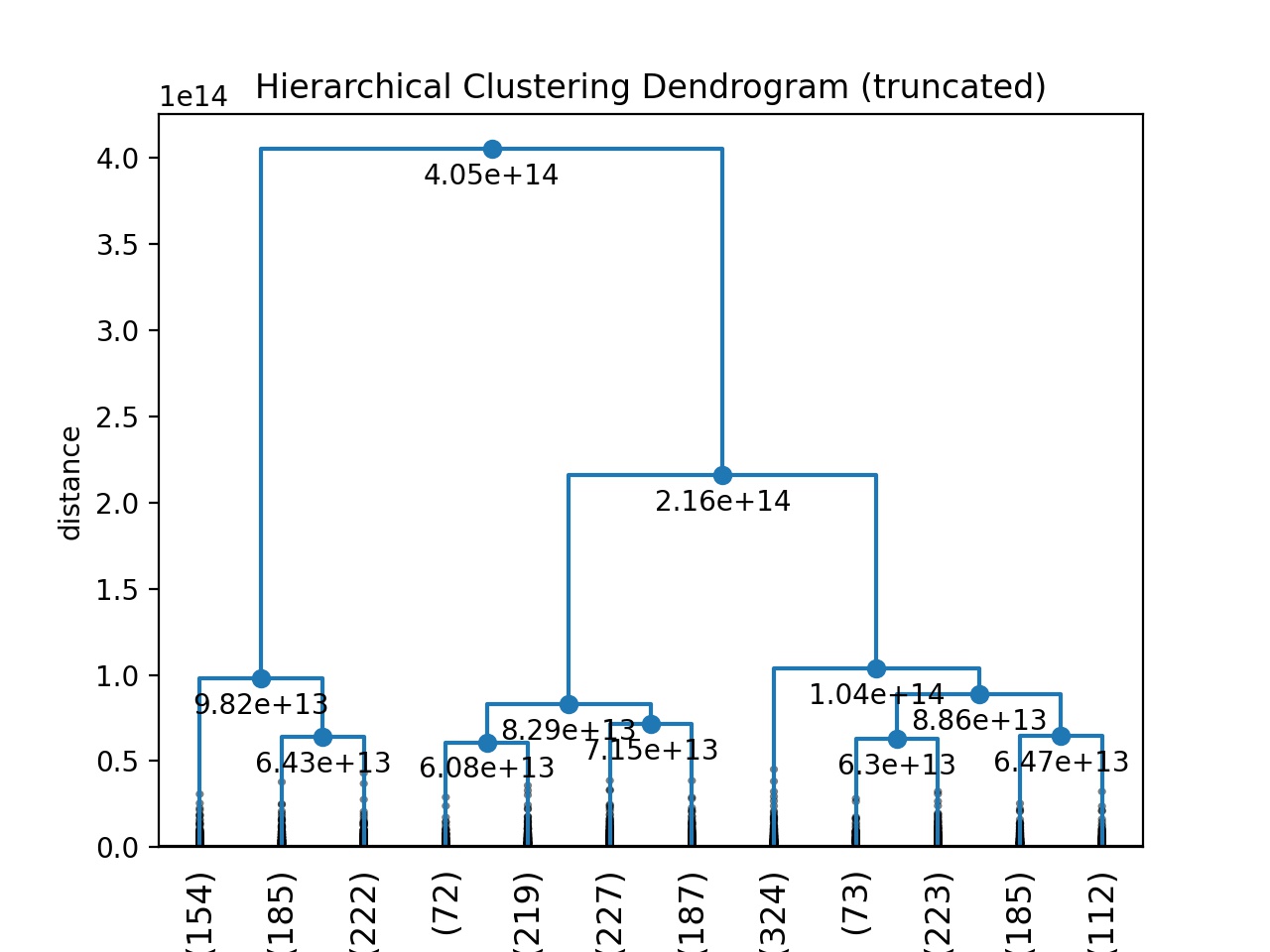
Create dendrogram.
Explain and interpret your results properly.
Examples¶
I prepare a python script for your reference based on Kretschmer et al. (2018).
Import functions and pachages
# ================================================================
# Yu-Chiao @ WHOI Feb 22, 2019
# Heirarchical cluster deomstration
# ================================================================
# ================================================================
# Import functions
# ================================================================
import argparse
from netCDF4 import Dataset
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import mlab
import sys, os, ast
import matplotlib.path as mpath
import matplotlib as mpl
from pylab import setp
import cartopy.crs as ccrs
import cartopy.feature as cfeature
from cartopy.mpl.ticker import LongitudeFormatter, LatitudeFormatter
import cartopy.io.img_tiles as cimgt
from cartopy.io.img_tiles import StamenTerrain
from scipy.cluster.hierarchy import dendrogram, linkage
from scipy.cluster.hierarchy import cophenet
from scipy.spatial.distance import pdist
from scipy.cluster.hierarchy import fcluster
Define functions
def fancy_dendrogram(*args, **kwargs):
max_d = kwargs.pop('max_d', None)
if max_d and 'color_threshold' not in kwargs:
kwargs['color_threshold'] = max_d
annotate_above = kwargs.pop('annotate_above', 0)
ddata = dendrogram(*args, **kwargs)
if not kwargs.get('no_plot', False):
plt.title('Hierarchical Clustering Dendrogram (truncated)')
plt.xlabel('sample index or (cluster size)')
plt.ylabel('distance')
for i, d, c in zip(ddata['icoord'], ddata['dcoord'], ddata['color_list']):
x = 0.5 * sum(i[1:3])
y = d[1]
if y > annotate_above:
plt.plot(x, y, 'o', c=c)
plt.annotate("%.3g" % y, (x, y), xytext=(0, -5),
textcoords='offset points',
va='top', ha='center')
if max_d:
plt.axhline(y=max_d, c='k')
return ddata
def reverse_colourmap(cmap, name = 'my_cmap_r'):
reverse = []
k = []
for key in cmap._segmentdata:
k.append(key)
channel = cmap._segmentdata[key]
data = []
for t in channel:
data.append((1-t[0],t[2],t[1]))
reverse.append(sorted(data))
LinearL = dict(zip(k,reverse))
my_cmap_r = mpl.colors.LinearSegmentedColormap(name, LinearL)
return my_cmap_r
def read_fields_here(filename):
f = Dataset(filename, 'r')
lon = f.variables['lon'][:].data
lat = f.variables['lat'][:].data
u10 = f.variables['var_in'][:,:,:].data
z10 = f.variables['var2_in'][:,:,:].data
f.close()
return lon, lat, u10, z10
Main script starts
# ================================================================
# main starts
# ================================================================
def main(inargs,year_st,year_ed):
"""Run the program."""
print('program finishes here')
# ================================================================
# Describe this script and control flags
# ================================================================
if __name__ == '__main__':
description='code template'
parser = argparse.ArgumentParser(description=description)
parser.add_argument("run_flag", type=str, help="False: do not run main program, just plotting figure; True: run the main program")
args = parser.parse_args()
plt.close('all')
flag_rm_tropics = 1
flag_Plumb = 0
flag_long_term_basic_flow = 1
np.set_printoptions(precision=5, suppress=True)
plt.close('all')
year = np.linspace(1979,2015,37)
Set cluster parameters and linking matrix
# ================================================================
# Set parameters
# ================================================================
# Set the number of clusters
k = 7
if ast.literal_eval(args.run_flag) == True:
filename = 'python_output_for_plot_tmp.nc'
[lon, lat, u10, z10] = read_fields_here(filename)
nt = u10.shape[0]
ny = u10.shape[1]
nx = u10.shape[2]
# ================================================================
# Perform hierarchical clustering
# ================================================================
# Reshape data
data_in = u10.reshape((nt,ny*nx)).copy()
# generate the linkage matrix
Z = linkage(data_in, 'ward')
plt.figure()
fancy_dendrogram(Z,truncate_mode='lastp',p=12,leaf_rotation=90.,leaf_font_size=12.,show_contracted=True,annotate_above=10, max_d=50)
plt.savefig('u_dendrogram_tmp_plot.jpg', format='jpeg', dpi=200)
# [c, coph_dists] = cophenet(Z, pdist(data_in))
# HERE is you can use different clusters
# Retrive clusters
clusters = fcluster(Z, k, criterion='maxclust')
# Construct time series
ts_u10 = np.zeros((k,int(nt/59)))
for NY in range(int(nt/59)):
for NN in range(k):
ts_u10[NN,NY] = sum(clusters[NY*59:NY*59+59]==NN+1)
ts_u10 = ts_u10/59.
percent_cluster = np.zeros((k))
for KK in range(k):
percent_cluster[KK] = np.sum(clusters==(KK+1))/len(clusters)*100.
z10_cluster = np.zeros((k,ny,nx))
for KK in range(k):
z10_cluster[KK,:,:] = np.nanmean(z10[clusters==(KK+1),:,:], axis=0)
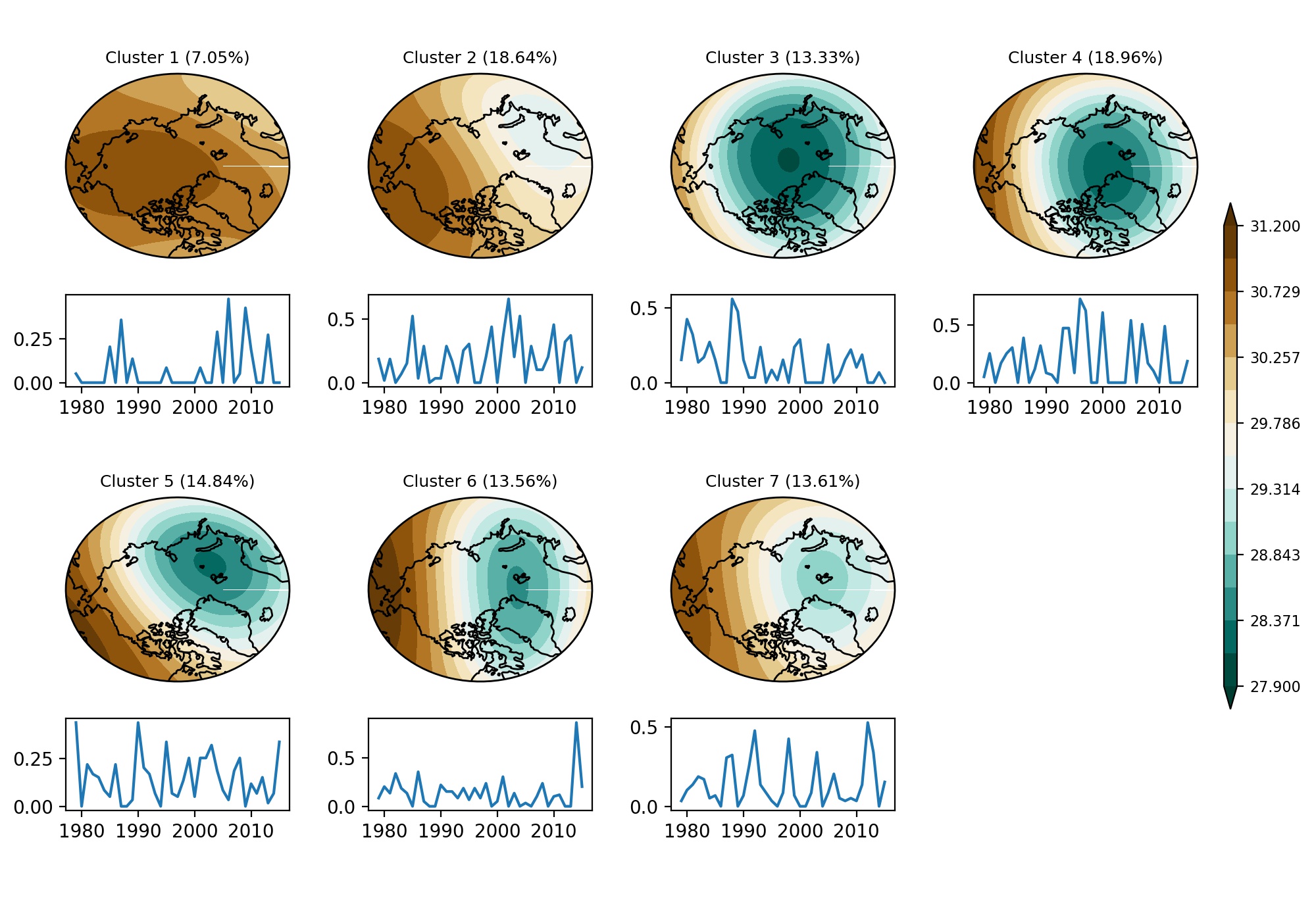
Plot figures
# ================================================================
# Plot figure
# ================================================================
clevel = np.linspace(27.9,31.2,15)
cmap = reverse_colourmap(plt.cm.BrBG)
font_size = 9
theta = np.linspace(0, 2*np.pi, 100)
center, radius = [0.5, 0.5], 0.5
verts = np.vstack([np.sin(theta), np.cos(theta)]).T
circle = mpath.Path(verts * radius + center)
fig = plt.figure()
fig.set_size_inches(10, 7, forward=True)
factor = 1.
for II in range(4):
ax = fig.add_axes([0.05+0.23*II, 0.72, 0.17, 0.2], projection=ccrs.Orthographic(-90, 90))
contourf = ax.contourf(lon, lat, z10_cluster[II,:,:], transform=ccrs.PlateCarree(), levels=clevel, cmap=cmap, extend='both')
ax.coastlines('110m')
ax.set_boundary(circle, transform=ax.transAxes)
ax.set_aspect('auto')
plt.title('Cluster ' + str(II+1) + ' (' + str(round(percent_cluster[II],2)) + '%)', fontsize=font_size)
ax = fig.add_axes([0.05+0.23*II, 0.58, 0.17, 0.1])
ax.plot(year, ts_u10[II,:])
for II in range(3):
ax = fig.add_axes([0.05+0.23*II, 0.26, 0.17, 0.2], projection=ccrs.Orthographic(-90, 90))
contourf = ax.contourf(lon, lat, z10_cluster[II+4,:,:], transform=ccrs.PlateCarree(), levels=clevel, cmap=cmap, extend='both')
ax.coastlines('110m')
ax.set_boundary(circle, transform=ax.transAxes)
ax.set_aspect('auto')
plt.title('Cluster ' + str(II+5) + ' (' + str(round(percent_cluster[II+4],2)) + '%)', fontsize=font_size)
ax = fig.add_axes([0.05+0.23*II, 0.12, 0.17, 0.1])
ax.plot(year, ts_u10[II+4,:])